3D Fibrin Network (interlacing tubular structures) Segmentation and Structure Identification
Fibrin network is an essential strucutre during the formation of blood clots. Quantitive analysis of the topological and morphological strucutres of the networks plays a critical role in the study of thrombus.
In fact, the common role of a computer algorithm plays is to reduce the human effort. But, in this problem, it is difficult even for human experts to manually examine and analyze the images due to the nature of 3D densely interlacing stuctures. So, an automated algorithm is of great importance.
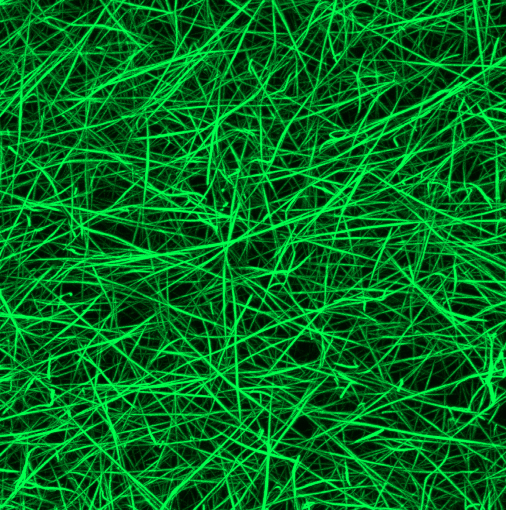
A Maximal Intensity Projection (MIP) visualization of a z-stack images of fibrin networks is shown below.
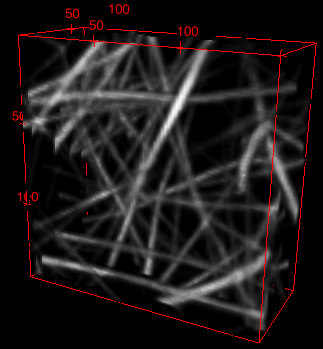
Here is the 3D reconstruction of a small region of the whole network.
The problem consists of two parts: obtaining the binary mask of fibrin networks from fluorescent images and identifying the network structure from the binary mask. (This is a typical step-by-step solution with analyzing the network structure as the ultimate goal. Other end-to-end models may be available.)
To do the segmentation, I used to a simple, but very effective strategy. The core idea is to apply a tubular structure enhancement/detection filter to locate where the fibrin fibers are. Then, a local thresholding is applied in each local window containing considerable amount of candidates.
The identification of the network structure is done by 3D thinning and structure prunning. First of all, morphological thinning is performed on the binary mask, which results in the one-voxel wide network skeletons.
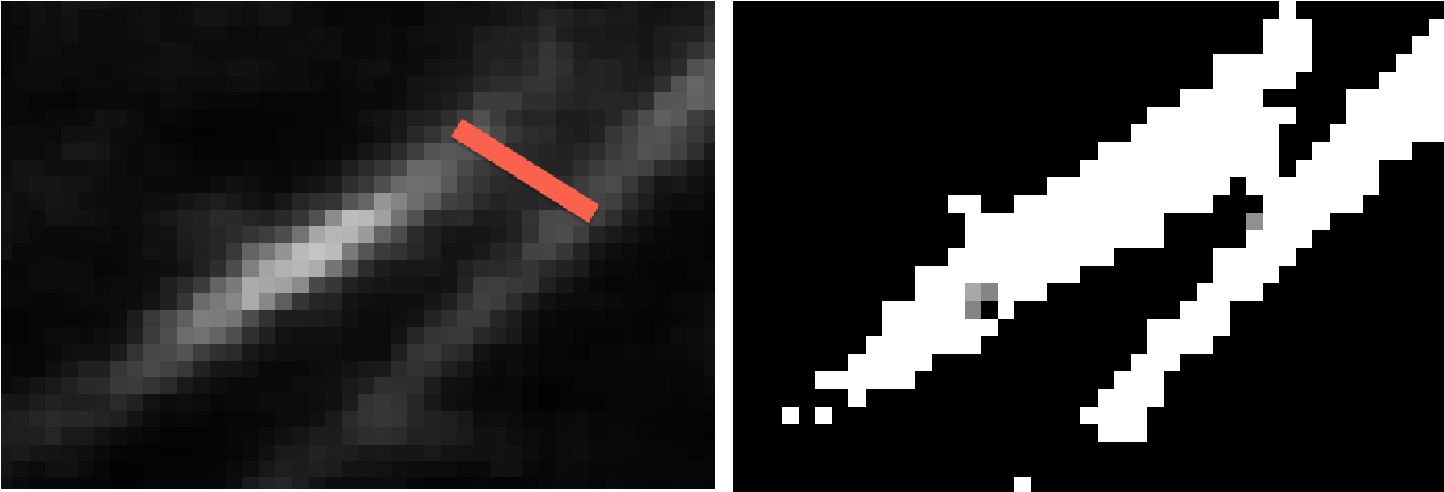
Second, we need to prune the short spurs caused by non-smoothness in the binary mask, false connections occur between proximal fibers, like the following case.
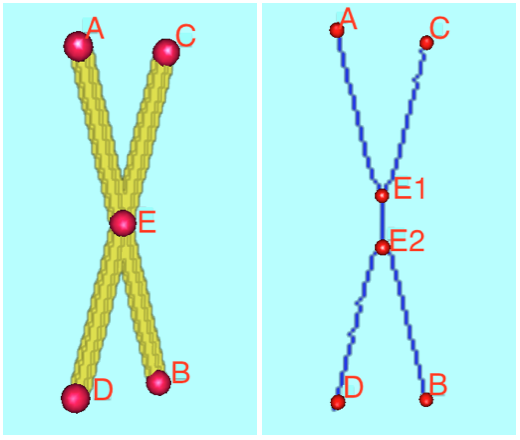
Finally, a common issue in 3D thinning is the duplication of intersection points. See the picutre below. We address this issue by analyzing the relative distance and angles between the connected branches of each pair of neighboring and proximal intersection points.
Evaluated on real 3D fibrin network datasets, our algorithm achieved signification improvement over the beest known method.
The source codes and more information can be found at here.